Sweet Regulation of Human Glucocorticoid Receptor Transcriptional Activity
Xin wen, Wen Wei Zeng
DOI10.21767/2471-8203.100004
1(V) Developmental Neurobiology Section, NHLBI, NIH, Bethesda, MD 20892, USA
2(C ) UGO, Eunice Kennedy Shriver, NICHD, NIH, Bethesda, MD 20892, USA
3University of Maryland, College Park, MD 20742, USA
- *Corresponding Author:
- Xin WEN
Developmental Neurobiology Section, NHLBI, NIH, Bethesda, MD 20892, USA
Tel: +12405063095
E-mail: xin.wen@nih.gov
Received date: August 19, 2015; Accepted date: September 15, 2015; Published date: September 22, 2015
Citation: WEN X (2015) Sweet Regulation of Human Glucocorticoid Receptor Transcriptional Activity. J Obes Eat Disord 1:9. doi: 10.4172/2471-8203.100004
Copyright: © 2016, Xin WEN. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
This study is to understanding the transcription profile of human Glucocorticoid Receptor GR, gene of a neuroendocrine, stress response and obesity related receptor. For GR, relative luciferase activity RLA and the distribution of phosphorylation [P], glycosylation sites [G] on transcription factor for promoter model were plotted on the same chart. Within the 3.2 kb upstream of the methionine ATG, for GR, trend lines of RLA with that of [G] or ([P]-[G]), both tend to have negative reciprocal relationship. It brings up the question: for the neuendocrine glucocorticoid receptor, does the nutrition and obesity related glycosylation regulated the transcriptional activity in a negative reciprocal way? In conclusion, for GR, the reciprocal relation between trend line of [G], ([P] ± [G]) or [P] and that of RLA, give a specific digit evidence for the first time to the theory, which the structure related glycosylation and the signal sensing phosphorylation, exhibit either independently or Interactively on regulation of transcription activity. Abbreviations: RLA: Relative Luciferase Activity; Luciferase/β-Galactosidase Activity; DLA: Dual Luciferase Assay; GR: Human Glucocorticoid Receptor; TF: Transcription Factor; P1: Construct Plasmid 1; P7: Construct Plasmid 7. GR-P1: Construct Plasmid 1 for Human Glucocorticoid Receptor; GR-P7: Constructs Plasmid 7 for Human Glucocorticoid Receptor; [P]: Number of Phosphorylation Sites on TF for Promoter Models; [G]: Number of Glycosylation Sites on TF for Promoter Models; [P]-[G]: Number of Difference between Phosphorylation and Glycosylation Sites on TF for Promoter Models; [P]+[G]: Number for Sum of Phosphoylation and Glycosylation Sites on TF for Promoter Models
Introduction
Glucocorticoid receptor signaling is in physiological and pathophysiological conditions in the major organ systems in the human body. Among the non-genomic signaling of GRs [1], the rapid actions of GRs, occur as a result of physiochemical interactions of glucocorticoids with the cell membrane; have been reported in various systems, including the cardiovascular, immune and neuroendocrine
In the embryonic development, the present of functional GR during gestation is essential for postnatal survival as well as during development. In the nervous system, GR functions in the brain correlate positively with anxiety behavior. The GR in the forebrain has been shown to regulate the HPA axis and behavior under stressed conditions. In the cardiovascular system, glucocorticoid regulation of cell size, apoptosis, inflammatory state, and vascular tone appears to be vital for proper cardiac function. GR signal is associated with immune system, respiratory system as well as reproductive system. Considerable evidence implicating GR signaling is in maintaining glucose homeostasis, regulating metabolic homeostasis e.g. Cushing’s disease or Addison’s disease. In musculoskeletal system, the activity of GR in skeletal muscle has been shown to correlate positively with metabolic syndrome. And in Integumentary system, the antiproliferative effects of the GR in keratinocytes were shown to be regulated by transrepression, and GR in the skin physiologically regulate epithelial integrity and immune function. And the glucocorticoid receptor plays the dominant role in adipogenesis and adipokine production in human adipocytes [2], On the other hand, primary gene induction or repression in eukaryotes does not require protein synthesis [3], suggesting the involvement of posttranslational modifications [4,5]. Since many different types of stimuli that affect gene expression also lead to the activation of protein kinases, analysis of transcription factor phosphorylation is essential for complete understanding of the signal pathways. The activity of transcription factors may be modulated by their signal-sensing domain including phosphorylation [6]. In addition, as nutrient sensitive sugar modification, glycosylation, interfere with the epigenetic control of gene expression.
GR receptors are involved in anxiety, cardiovascular, apoptosis, inflammatory, immune system, respiratory, reproductive system, musculoskeletal and integumentary system diseases, as well as obesity and neuroendocrine disease. Moreover, phosphorylation or glycosylation are perhaps required for the activation of transcription factors. Thus, the regulatory mechanism and post translation modification in the epigenetic transcription regulation of glucocorticoid receptor would be beneficial.
Methods
For GR, relative luciferase activity (in HeLa (human cervical carcinoma cells) is from Figure 7 in Vedeckis’s paper [7], and the sequence information for the corresponding constructs is from Figure 4 in the same paper. Afterwards, promoter model [8] [A promoter model represents a framework of two or more conserved elements (e.g., transcription factor binding sites) with a defined distance (and strand orientation). Usually, promoter models are much more specific than single elements like transcription factor binding sites. Therefore, a promoter model can give higher evidence that the matching sites are functional was inspected by Genomatix (https://www.genomatix.com/). The glycosylation (Table 1) and phosphorylation sites (Table 2) on the promoter model were searched by Protein Knowledge bases of Uniprot (https://www.uniprot.org/).
| Promoter Model | Sequence | bp | G1 | TF |
|---|---|---|---|---|
| EGRF_SP1F_01 | GTTGGGGGCGGGGGGCG | -3096 | G | V$SP1F |
| SP1F_SP1F_01 | GTTGGGGGCGGGGGGCG | -3096 | G | V$SP1F |
| SP1F_SP1F_06 | GTTGGGGGCGGGGGGCG | -3096 | G | V$SP1F |
| SP1F_ETSF_04 | GTTGGGGGCGGGGGGCG | -3096 | G | V$SP1F |
| SP1F_KLFS_01 | GTTGGGGGCGGGGGGCG | -3096 | G | V$SP1F |
| SP1F_SP1F_06 | GCGGGGGGCGAAGCGCG | -3089 | G | V$SP1F |
| SP1F_SP1F_01 | GCGGGGGGCGAAGCGCG | -3089 | G | V$SP1F |
| SP1F_SP1F_05 | GCGGGGGGCGAAGCGCG | -3089 | G | V$SP1F |
| SP1F_SP1F_01 | GCACCGGGCGGGGCGGC | -3070 | G | V$SP1F |
| KLFS_SP1F_01 | GCACCGGGCGGGGCGGC | -3070 | G | V$SP1F |
| SP1F_SP1F_06 | GCACCGGGCGGGGCGGC | -3070 | G | V$SP1F |
| SP1F_KLFS_01 | GCACCGGGCGGGGCGGC | -3070 | G | V$SP1F |
| SP1F_SP1F_05 | GCACCGGGCGGGGCGGC | -3070 | G | V$SP1F |
| SP1F_SP1F_01 | GGGCGGGGCGGCCACGC | -3065 | G | V$SP1F |
| SP1F_SP1F_06 | GGGCGGGGCGGCCACGC | -3065 | G | V$SP1F |
| SP1F_SP1F_01 | CGGGGTGGCGGGGCCCG | -3015 | G | V$SP1F |
| E2FF_SP1F_01 | GCGGAGGGCGTGGGGGC | -2997 | G | V$SP1F |
| SP1F_NF1F_01 | GCGGAGGGCGTGGGGGC | -2997 | G | V$SP1F |
| SP1F_SP1F_01 | CGTGGGGGCAGGGACCG | -2989 | G | V$SP1F |
| SP1F_E2FF_01 | CCCTCGGGCGGGGAGCG | -2906 | G | V$SP1F |
| SP1F_EBOX_SP1F_01 | CCCTCGGGCGGGGAGCG | -2906 | G | V$SP1F |
| SP1F_EBOX_SP1F_01 | CCCTCGGGCGGGGAGCG | -2906 | G | V$SP1F |
| E2FF_SP1F_01 | GCCGGGGGTGGAGTGGG | -2889 | G | V$SP1F |
| EBOX_EBOX_02 | GGAGCGCGTGTGT | -2876 | G | V$EBOX |
| EBOX_EBOX_02 | GCGCCACGGCGCG | -2852 | G | V$EBOX |
| SP1F_EBOX_SP1F_01 | GCGCCACGGCGCG | -2852 | G | V$EBOX |
| SP1F_EBOX_SP1F_01 | GCGCCACGGCGCG | -2852 | G | V$EBOX |
| SP1F_EBOX_SP1F_01 | CGAGCGAGCGGGACCGA | -2817 | G | V$SP1F |
| SP1F_EBOX_SP1F_01 | GGCCTGGGCGAGCGAGC | -2809 | G | V$SP1F |
| P12 | -2738 | |||
| SP1F_ETSF_02 | GCGCGGGGCGGAGGGCT | -2584 | G | V$SP1F |
| SP1F_ETSF_03 | GCGCGGGGCGGAGGGCT | -2584 | G | V$SP1F |
| SP1F_ETSF_03 | TCCATGGGTGGGGGGAG | -2524 | G | V$SP1F |
| EBOX_E2FF_01 | CCGCCACCGTCCG | -2400 | G | V$EBOX |
| ETSF_SP1F_01 | TCCGCAGGCGTCCCCTG | -2164 | G | V$SP1F |
| ETSF_SP1F_05 | TGGCCGGGCCGAGGGGG | -2149 | G | V$SP1F |
| P2 | -1824 | |||
| SP1F_SP1F_01 | GGCCGGGGCCGGCGTTA | -1810 | G | V$SP1F |
| SP1F_SP1F_01 | GAAGTGGGCGTGTCGGA | -1786 | G | V$SP1F |
| SP1F_KLFS_01 | TTGCGGGGCGGGGGTGG | -1710 | G | V$SP1F |
| EGRF_SP1F_01 | TTGCGGGGCGGGGGTGG | -1710 | G | V$SP1F |
| P3 | -1630 | |||
| P4 | -1525 | |||
| NKXH_CEBP_01 | TCCCTCAAGCGACATTATC | -1457 | G | V$NKXH |
| NFAT_SORY_01 | CCAAAACAATATTTCCTAAAACGAA | -1430 | G | V$SORY |
| SORY_SORY_01 | CCAAAACAATATTTCCTAAAACGAA | -1430 | G | V$SORY |
| CREB_IRFF_01 | CTTTTTTGACAGCTGCCTTCA | -1398 | G | V$CREB |
| SORY_SORY_01 | CCAATGAATTTCCATGCCGCTTTTT | -1381 | G | V$SORY |
| P5 | -1322 | |||
| SP1F_KLFS_01 | GAGAGGGGTGTGGACTT | -1260 | G | V$SP1F |
| CREB_NFKB_05 | ATGCGATGACGTTAGGCAGCA | -1198 | G | V$CREB |
| P6 | -1149 | |||
| P7 | -1115 | |||
| NEUR_SORY_01 | AATGAATTATAATGTCTGTGATTAA | -324 | G | V$SORY |
1. G: glycosylation site.
2. P1: Plasmid Construct 1.
Table 1: Glycosylation site on promoter model of GR.
| Promoter Model | Sequence | bp | P1 | TF |
|---|---|---|---|---|
| SP1F_ETSF_04 | ACTCCCCAGGAAAAAGGGTGG | -3113 | P | V$ETSF |
| EGRF_SP1F_01 | GGAGTTGGGGGCGGGGG | -3099 | P | V$EGRF |
| SP1F_KLFS_01 | AGTTGGGGGCGGGGGGC | -3097 | P | V$KLFS |
| EGRF_SP1F_01 | GTTGGGGGCGGGGGGCG | -3096 | P | V$SP1F |
| SP1F_SP1F_01 | GTTGGGGGCGGGGGGCG | -3096 | P | V$SP1F |
| SP1F_SP1F_06 | GTTGGGGGCGGGGGGCG | -3096 | P | V$SP1F |
| SP1F_ETSF_04 | GTTGGGGGCGGGGGGCG | -3096 | P | V$SP1F |
| SP1F_KLFS_01 | GTTGGGGGCGGGGGGCG | -3096 | P | V$SP1F |
| KLFS_SP1F_01 | GGCGGGGGGCGAAGCGC | -3090 | P | V$KLFS |
| SP1F_SP1F_06 | GCGGGGGGCGAAGCGCG | -3089 | P | V$SP1F |
| SP1F_SP1F_01 | GCGGGGGGCGAAGCGCG | -3089 | P | V$SP1F |
| SP1F_SP1F_05 | GCGGGGGGCGAAGCGCG | -3089 | P | V$SP1F |
| SP1F_KLFS_01 | CGCACCGGGCGGGGCGG | -3071 | P | V$KLFS |
| SP1F_SP1F_01 | GCACCGGGCGGGGCGGC | -3070 | P | V$SP1F |
| KLFS_SP1F_01 | GCACCGGGCGGGGCGGC | -3070 | P | V$SP1F |
| SP1F_SP1F_06 | GCACCGGGCGGGGCGGC | -3070 | P | V$SP1F |
| SP1F_KLFS_01 | GCACCGGGCGGGGCGGC | -3070 | P | V$SP1F |
| SP1F_SP1F_05 | GCACCGGGCGGGGCGGC | -3070 | P | V$SP1F |
| SP1F_SP1F_01 | GGGCGGGGCGGCCACGC | -3065 | P | V$SP1F |
| SP1F_SP1F_06 | GGGCGGGGCGGCCACGC | -3065 | P | V$SP1F |
| SP1F_SP1F_01 | CGGGGTGGCGGGGCCCG | -3015 | P | V$SP1F |
| E2FF_SP1F_01 | GGGTGGCGGGGCCCGCG | -3013 | P | V$E2FF |
| EBOX_E2FF_01 | TCCGCGCGGGCCCCGCC | -3009 | P | V$E2FF |
| SP1F_NF1F_01 | GCGGAGGGCGTGGGGGC | -2997 | P | V$SP1F |
| SP1F_SP1F_01 | CGTGGGGGCAGGGACCG | -2989 | P | V$SP1F |
| SP1F_NF1F_01 | CGCCCCTGCAGTTGCCAAGCG | -2966 | P | V$NF1F |
| IKRS_AP2F_01 | CGCGGGGAACGAT | -2934 | P | V$IKRS |
| SP1F_E2FF_01 | GCGCGGCGGCCGCGGGG | -2926 | P | V$E2FF |
| IKRS_AP2F_01 | CCCGCCCGAGGGGCC | -2910 | P | V$AP2F |
| SP1F_E2FF_01 | CCCTCGGGCGGGGAGCG | -2906 | P | V$SP1F |
| SP1F_EBOX_SP1F_01 | CCCTCGGGCGGGGAGCG | -2906 | P | V$SP1F |
| SP1F_EBOX_SP1F_01 | CCCTCGGGCGGGGAGCG | -2906 | P | V$SP1F |
| E2FF_SP1F_01 | CTCGGGCGGGGAGCGGC | -2904 | P | V$E2FF |
| E2FF_SP1F_01 | GCCGGGGGTGGAGTGGG | -2889 | P | V$SP1F |
| EBOX_EBOX_02 | GGAGCGCGTGTGT | -2876 | P | V$EBOX |
| EBOX_EBOX_02 | GCGCCACGGCGCG | -2852 | P | V$EBOX |
| SP1F_EBOX_SP1F_01 | GCGCCACGGCGCG | -2852 | P | V$EBOX |
| SP1F_EBOX_SP1F_01 | GCGCCACGGCGCG | -2852 | P | V$EBOX |
| SP1F_EBOX_SP1F_01 | CGAGCGAGCGGGACCGA | -2817 | P | V$SP1F |
| SP1F_EBOX_SP1F_01 | GGCCTGGGCGAGCGAGC | -2809 | P | V$SP1F |
| P12 | ||||
| AP1R_ETSF_EGRF_01 | GATTCTGTGGGTGGAAG | -2682 | P | V$EGRF |
| AP1R_ETSF_EGRF_01 | CTGTGGGTGGAAGGAGACGCC | -2676 | P | V$ETSF |
| AP1R_ETSF_EGRF_01 | AGCTGCTTCGGCCGCTCCGGC | -2652 | P | V$AP1r |
| SP1F_ETSF_03 | GCGCGCCCGGAACCTCGACCC | -2613 | P | V$ETSF |
| SP1F_ETSF_02 | GCGCGGGGCGGAGGGCT | -2584 | P | V$SP1F |
| SP1F_ETSF_03 | GCGCGGGGCGGAGGGCT | -2584 | P | V$SP1F |
| STAT_NFKB_06 | GGGGGAGAGCCCCTA | -2533 | P | V$NFKB |
| 1F_ETSF_03 | TCCATGGGTGGGGGGAG | -2524 | P | V$SP1F |
| SP1F_ETSF_03 | TAAAAAAAGGAAGTAAACAGC | -2495 | P | V$ETSF |
| STAT_NFKB_06 | TTTTTTCTAAAAAAAGGAA | -2487 | P | V$STAT |
| IRFF_NFAT_01 | TTTAGAAAAAAAAAATATATT | -2478 | P | V$IRFF |
| IRFF_NFAT_01 | AGGAGGGAAATATATTTTT | -2469 | P | V$NFAT |
| SMAD_E2FF_01 | CTCGGCTGCGG | -2421 | P | V$SMAD |
| SMAD_E2FF_01 | CTGCGGCGGGAACTGCG | -2413 | P | V$E2FF |
| SP1F_ETSF_02 | CTGCGGCGGGAACTGCGGACG | -2411 | P | V$ETSF |
| EBOX_E2FF_01 | CCGCCACCGTCCG | -2400 | P | V$EBOX |
| AP2F_KLFS_01 | ACTCCCCGAGGCTAA | -2261 | P | V$AP2F |
| AP2F_KLFS_01 | GCCTCGGGGAGTGGGGG | -2257 | P | V$KLFS |
| ETSF_SP1F_01 | GAGGGAGAGGAAGAGGCCAGC | -2181 | P | V$ETSF |
| Promoter Model | Sequence | bp | P | TF |
| ETSF_SP1F_01 | TCCGCAGGCGTCCCCTG | -2164 | P | V$SP1F |
| ETSF_SP1F_05 | TGGCCGGGCCGAGGGGG | -2149 | P | V$SP1F |
| ETSF_SP1F_05 | GAGGGGGAGGAACCTGACCTC | -2137 | P | V$ETSF |
| P2 | -1824 | |||
| SP1F_SP1F_01 | GGCCGGGGCCGGCGTTA | -1810 | P | V$SP1F |
| SP1F_SP1F_01 | GAAGTGGGCGTGTCGGA | -1786 | P | V$SP1F |
| SP1F_KLFS_01 | TTGCGGGGCGGGGGTGG | -1710 | P | V$SP1F |
| EGRF_SP1F_01 | TTGCGGGGCGGGGGTGG | -1710 | P | V$SP1F |
| SP1F_KLFS_01 | CTTGCGGGGCGGGGGTG | -1709 | P | V$KLFS |
| EGRF_SP1F_01 | CCCTTGCGGGGCGGGGG | -1707 | P | V$EGRF |
| P3 | -1630 | |||
| BRNF_RXRF_01 | GTGGTATTACAAGGTTGCA | -1545 | P | V$BRNF |
| P4 | -1525 | |||
| BRNF_RXRF_01 | TGGCATGGTTCATTAGGGCCAATTA | -1512 | P | V$RXRF |
| NKXH_CEBP_01 | TCCCTCAAGCGACATTATC | -1457 | P | V$NKXH |
| NKXH_CEBP_01 | CGTTTTAGGAAATAT | -1433 | P | V$CEBP |
| NFAT_SORY_01 | CCAAAACAATATTTCCTAAAACGAA | -1430 | P | V$SORY |
| SORY_SORY_01 | CCAAAACAATATTTCCTAAAACGAA | -1430 | P | V$SORY |
| NFAT_SORY_01 | TTTTAGGAAATATTGTTTT | -1429 | P | V$NFAT |
| IRFF_NFAT_01 | TTTTAGGAAATATTGTTTT | -1429 | P | V$NFAT |
| IRFF_NFAT_01 | ACCCGAAACCAAAACAATATT | -1420 | P | V$IRFF |
| CREB_IRFF_01 | CTTCAAACCCGAAACCAAAAC | -1414 | P | V$IRFF |
| PBXC_MYOD_01 | TTTTGACAGCTGCCTTC | -1399 | P | V$MYOD |
| CREB_IRFF_01 | CTTTTTTGACAGCTGCCTTCA | -1398 | P | V$CREB |
| PBXC_MYOD_01 | CGCTTTTTTGACAGCTG | -1394 | P | V$PBXC |
| TEAF_TEAF_01 | TTCCATGCCGCTT | -1384 | P | V$TEAF |
| SORY_SORY_01 | CCAATGAATTTCCATGCCGCTTTTT | -1381 | P | V$SORY |
| NFAT_GATA_01 | CATGGAAATTCATTGGGCT | -1375 | P | V$NFAT |
| TEAF_TEAF_01 | CTCCATTCGATAC | -1361 | P | V$TEAF |
| NFAT_GATA_01 | AGGCGATAACGAT | -1335 | P | V$GATA |
| PAX6_CDXF_01 | TGCCCCGTTTATCTGAGGC | -1323 | P | V$CDXF |
| P5 | -1322 | |||
| CREB_NFKB_05 | TGTGGACTTGCCACT | -1267 | P | V$NFKB |
| SP1F_KLFS_01 | GAGAGGGGTGTGGACTT | -1260 | P | V$SP1F |
| SP1F_KLFS_01 | AGAGAGGGGTGTGGACT | -1259 | P | V$KLFS |
| CREB_NFKB_05 | ATGCGATGACGTTAGGCAGCA | -1198 | P | V$CREB |
| P6 | -1149 | |||
| P7 | -1115 | |||
| FKHD_CEBP_01 | CCTTTCCAAACAAATAT | -913 | P | V$FKHD |
| FKHD_CEBP_01 | TTTGTTTGGAAAGGA | -911 | P | V$CEBP |
| NFKB_CEBP_01 | TTTGTTTGGAAAGGA | -911 | P | V$CEBP |
| NFKB_CEBP_01 | TAAGTTCTTTCCTTT | -902 | P | V$NFKB |
| STAT_BRAC_01 | TTGGTTCTCAGAAAAGCAA | -448 | P | V$STAT |
| PBXC_PDX1_01 | GCCGTGATTGAAAAGAG | -427 | P | V$PBXC |
| PBXC_PDX1_01 | TAGGAATTTTAATGATCAC | -408 | P | V$PDX1 |
| STAT_BRAC_01 | AAAAAAAGGAAGTGTGATCAT | -396 | P | V$BRAC |
| BRNF_RXRF_01 | TGAAGGTTCAAGTTGATGTCAAAGT | -363 | P | V$RXRF |
| NEUR_SORY_01 | ATTACATCTGATT | -340 | P | V$NEUR |
| BRNF_RXRF_01 | ATGTAATGAATTATAATGT | -331 | P | V$BRNF |
| NEUR_SORY_01 | AATGAATTATAATGTCTGTGATTAA | -324 | P | V$SORY |
| SMAD_FKHD_01 | AATGTCTGTGA | -321 | P | $SMAD |
| SMAD_FKHD_01 | CTGTGATTAACAAAGCT | -313 | P | V$FKHD |
| HNF1_CEBP_01 | TTTATTCTGGAAGAT | -170 | P | V$CEBP |
| HNF1_FKHD_01 | GATTCGGAGTTAACTAA | -124 | P | V$HNF1 |
| HNF1_CEBP_01 | GATTCGGAGTTAACTAA | -124 | P | V$HNF1 |
| HNF1_FKHD_01 | GTTCATTTAACAAGCTG | -104 | P | V$FKHD |
| GATA_HNF1_01 | GGCAGCTTGTTAAATGA | -102 | P | V$HNF1 |
| GATA_HNF1_01 | ATCCGATTAGTAA | -85 | P | V$GATA |
| ETSF_MYBL_01 | TCGGATCAGGAAGATAATGTG | -74 | P | V$ETSF |
| ETSF_MYBL_01 | CAAAAACGGGGGGAA | -36 | P | V$MYBL |
G: phosphorylation site.
P1: Plasmid Construct 1.
Table 2 Phosphorylation site on promoter model of GR.
Results
Human Glucocorticoid Receptor
As shown in Figure 1, GR-P1 has RLA of 100, with length of 1692bp, ranging from -2738bp to -1046bp, there are 41 phosphorylation sites and 4 glycosylaton sites on modules between GR-P1 and GR-P2. GR-P2 has RLA of 70.3, with length of 778bp, ranging from -1824bp to -1046bp, there are 22 phosphorylation sites and 6 glycosylaton sites on modules between GR-P2 and GR-P3. GR-P3 has RLA of 80.8, with length of 584bp, ranging from -1630bp to -1046bp, there are 6 phosphorylation sites and 4glycosylaton sites on modules between GR-P3 and GRP4. GR-P4 has RLA of 77.8, with length of 479bp, ranging from -1525bp to -1046bp, there are 1 phosphorylation sites and 0 glycosylaton sites onmodules between GR-P4 and GR-P5. GR-P5 has RLA of 55.9, with length of 276bp, ranging from -1322bp to -1046bp, there are 18 phosphorylation sites and 5 glycosylaton sites on modules between GR-P5 and GR-P6. GR-P6 has RLA of 1.8, with length of 103 bp, ranging from -1149 bp to -1046bp, there are 4 phosphorylation sites and 2 glycosylaton sites on modules between GR-P6 and GR-P7. GR-P7 has RLA of 2.5, with length of 69bp, ranging from -1115bp to -1046bp, there are 22 phosphorylation sites and 1 glycosylaton sites on modules on GR-P7.
Figure 1: RLA of corresponding plasmids for GR and distribution of phosphorylation site and glycosylation site on TF for promoter model. The RLA of the corresponding plasmids is plotted on upper Y axis (Y>0) with the corresponding sequence of sense primer and that of antisense primer, concurrently, a horizontal line is drawn to link the starting point and ending point of each constructed plasmid respectively. On the other hand, within the 3200 base pair (bp) upstream the methionineATG (0 bp), on the lower Y axis area (Y<0), the phosphorylation (P) sites and glycosylation (G) sites is also pinned on the corresponding sequence (bp) for transcription factors promoter models. It shows the distribution of P and G. GR, plasmids (P1, P2, P3, 21, P4, P5, P6, P7).
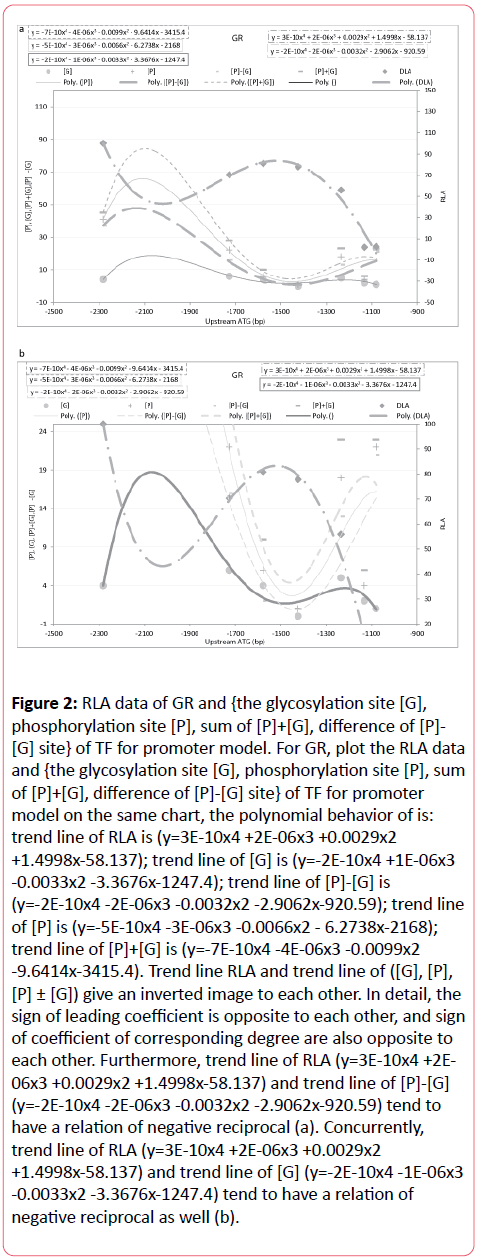
If plot according to the RLA data and {the glycosylation site [G], phosphorylation site [P], sum of [P]+[G], difference of [P]-[G] site} of TF for promoter model on the same chart, get Figure 2, the polynomial behavior (Table 3) of GR is: trend line of RLA is (y=3E-10x4+2E-06x3+0.0029x2+1.4998x-58.137); trend line of [G] is (y=2E-10x4-1E-06x3-0.0033x2-3.3676x-1247.4); trend line of [P]- [G] is (y=-2E-10x4-2E-06x3-0.0032x2-2.9062x-920.59); trend line of [P] is (y=-5E-10x4-3E-06x3-0.0066x2-6.2738x-2168); trend line of [P]+[G] is (y=-7E-10x4-4E-06x3-0.0099x2-9.6414x-3415.4). Trend line RLA and trend line of ([G], [P], [P] ± [G]) give an inverted image to each other. In detail, the sign of leading coefficient is opposite to each other, and sign of coefficient of corresponding degree are also opposite to each other. Furthermore, trend line of RLA (y=-2E- 10x4-1E-06x3-0.002x2-2.313x-546.1) and trend line of [G] (y=-2E- 10x4-1E-06x3-0.0033x2-3.3676x-1247.4) tend to have a relation of negative reciprocal (Figure 2b). At the same time, trend line of RLA (y=-2E-10x4-1E-06x3-0.002x2-122.313x-546.1) and trend line of [P]- [G] (y=-2E-10x4-2E-06x3-0.0032x2-2.9062x-920.59) tend to have a relation of negative reciprocal as well (Figure 2a).
| Y axis | polynomial scheme |
|---|---|
| RLA | y=3E-10x4+2E-06x3+0.0029x2+1.4998x-58.137 |
| [G] | y=-2E-10x4-1E-06x3-0.0033x2-3.3676x-1247.4 |
| [P]-[G] | y=-2E-10x4-2E-06x3-0.0032x2-2.9062x-920.59 |
| [P]+[G] | y=-7E-10x4-4E-06x3-0.0099x2-9.6414x-3415.4 |
Table 3: polynomial scheme of GR.
Figure 2: RLA data of GR and {the glycosylation site [G], phosphorylation site [P], sum of [P]+[G], difference of [P]- [G] site} of TF for promoter model. For GR, plot the RLA data and {the glycosylation site [G], phosphorylation site [P], sum of [P]+[G], difference of [P]-[G] site} of TF for promoter model on the same chart, the polynomial behavior of is: trend line of RLA is (y=3E-10x4 +2E-06x3 +0.0029x2 +1.4998x-58.137); trend line of [G] is (y=-2E-10x4 +1E-06x3 -0.0033x2 -3.3676x-1247.4); trend line of [P]-[G] is (y=-2E-10x4 -2E-06x3 -0.0032x2 -2.9062x-920.59); trend line of [P] is (y=-5E-10x4 -3E-06x3 -0.0066x2 - 6.2738x-2168); trend line of [P]+[G] is (y=-7E-10x4 -4E-06x3 -0.0099x2 -9.6414x-3415.4). Trend line RLA and trend line of ([G], [P], [P] ± [G]) give an inverted image to each other. In detail, the sign of leading coefficient is opposite to each other, and sign of coefficient of corresponding degree are also opposite to each other. Furthermore, trend line of RLA (y=3E-10x4 +2E- 06x3 +0.0029x2 +1.4998x-58.137) and trend line of [P]-[G] (y=-2E-10x4 -2E-06x3 -0.0032x2 -2.9062x-920.59) tend to have a relation of negative reciprocal (a). Concurrently, trend line of RLA (y=3E-10x4 +2E-06x3 +0.0029x2 +1.4998x-58.137) and trend line of [G] (y=-2E-10x4 -1E-06x3 -0.0033x2 -3.3676x-1247.4) tend to have a relation of negative reciprocal as well (b).
| EDI | PREmean (SD) | POSTmean (SD) | Diference(CI 95%) | p |
|---|---|---|---|---|
| Drive for thinness | 9.60 (5.21) | 6.88 (4.33) | -2.72 (-4.68; -0.76) | 0.008 |
| Bulimia | 3.04 (4.59) | 1.00 (2.66) | -2.04 (-3.94;-0.14) | 0.16 |
| Body Dissatisfaction | 19.20 (4.88) | 11.36 (7.11) | -7.84 (-11.24; -0.14) | 0.65 |
| Ineffectiveness | 4.88 (5.47) | 4.00 (6.00) | -0.88 (-2.70;0.94) | <0.001 |
| Perfectionism | 4.12 (2.99) | 3.36 (2.97) | -0.76 (-2.65; 1.13) | 0.38 |
| Interpersonal Distrust | 3.32 (3.98) | 2.88 (3.94) | -0.44 (-2.01; 1.13) | 0.006 |
| Interoceptive Awareness | 6.48 (4.98) | 5.28 (4.90) | -1.20 (-3.44; 1.04) | 0.05 |
| Maturity Fears | 4.92 (4.13) | 4.16 (3.36) | -0.76 (-2.83; 1.31) | 0.59 |
| BITE | ||||
| Symptoms | 12.04 (7.00) | 6.32 (4.25) | -5.72 (-8.13; -3.31) | 0.004 |
| Severity | 4.80 (4.94) | 2.16 (3.06) | -2.64 (-4.48; -0.80) | 0.021 |
| ROSEMBERG | 29.20 (7.10) | 32.64 (4.94) | 3.44 (0.21; 6.66) | 0.34 |
| BDI | 16.35 (10.36) | 11.69 (10.09) | -4.65 (-8.18; -1.12) | <0.001 |
| HDRS | 13.21 (6.88) | 5.58 (7.00) | -7.63 (-11.77;-3.50) | 0.33 |
| BSQ | 114.68 (38.62) | 92.40 (39.29) | -22.28 (-35.71;-8.85) | <0.001 |
Table 4 Differences in clinical variables before and after 1 year postsurgery in 22 females patients with morbid obesity.
Discussion
As a result, trend line of RLA (y=-2E-10x4-1E-06x3-0.002x2-2.313x-546.1) and trend line of [G] (y=-2E-10x4-1E-06x3-0.0033x2- 3.3676x-1247.4) tend to have a relation of negative reciprocal (Figure 2b). At the same time, trend line of RLA (y = -2E-10x4 - 1E-06x3 - 0.002x2 - 2.313x - 546.1) and trend line of [P]-[G] (y=-2E- 10x4-2E-06x3-0.0032x2-2.9062x-920.59) tend to have a relation of negative reciprocal as well (Figure 2a). Further investigations are being made to verify the reciprocal relationship.
From the fact shown by the figure that trend line of RLA of GR tends to have a relation of negative reciprocal to that the trend line of the corresponding ([P]-[G]) (Figure 2a), and trend line of ([P]- [G]) is the closest one (among the negative reciprocal relations) which approaching to its corresponding negative reciprocal RLA (Table 3). If it is true, the structure and nutritional element [G] will reduce the regulation of signal sensing phosphorylation [P] to the transcription activity; [P] and [G] coordinately play a negative reciprocal regulation to the transcription activity? Since O-GlcNAc and O-phosphate exhibit a complex interplay on signaling, transcriptional, and cytoskeletal regulatory proteins within the cell, one of the major functions of O-GlcNAc is to prevent O-phosphorylation and, by doing so, to modulate signaling and transcription in response to cellular nutrients or stress [9], does this negative reciprocal between trend line of ([P]- [G]) and that of RLA give a specific digit evidence to “O-GlcNAc prevent O-phosphorylation” in the above theory? Notice here is glycosylation, not O-GlcNAc.
In contrast, as indicated from the figure that the trend line of RLA tend to have a relation of negative reciprocal to that the trend line of [G] (Figure 2b) and trend line of [G] is the second closet one (among the negative reciprocal relations) to its negative reciprocal RLA (Table 3). If it is true, though Change of signal sensing phosphorylation [P] influence the negative reciprocal regulation of transcription activity by [G], [P] is related to [G], but [G] is independent from [P]. It helps us recall the theory: O-GlcNAc and O-phosphate exhibit a complex interplay on signaling, transcriptional, and cytoskeletal regulatory proteins within the cell, sometimes, O-GlcNAcylation and O-phosphorylation appear to be independently regulated. Does this figure give specific digit evidence to “independently regulated” in the above theory?
Moreover, for all three receptors, as shown in Table 3, sequence of the trend line is: |[G]|<|([P]-[G])|~|RLA|<|[P]|<|([P]+[G])|, that the ([P]+[G]) is the farthest away from the RLA. From opposite aspect, which gives another specific digit proof to the above theory, major function of O- GlcNAc is to prevent O-phosphorylation. Since if [G] play a positive role in the [P], trend line of the ([P]+[G]) would be closer to that of RLA.
Now the question of investigation is does the nutritional and obesity related glycosylation and the signal sensing phosphorylation regulated the transcriptional activity in a negative reciprocal way for the neroendocrine glucocorticoid receptors? Besides, no matter such negative reciprocal exist or not in experimental world, simply from the negative reciprocal relationship indicated by the RLA~([G] or [P]-[G]) figure, we can predict the transcriptional activity of the GR in the same cell type as shown in their corresponding RLA, by counting the [P] and [G].
Declaration of Interest
There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported
Funding
This research did not receive any specific grant from any funding agency in the public, commercial or not-for-profit sector.
References
- Kadmiel M, Cidlowski JA (2013) Glucocorticoid receptor signaling in health and disease. Trends PharmacolSci 24: 518-530.
- Lee MJ, Fried SK (2014) The glucocorticoid receptor, not the mineralocorticoid receptor, plays the dominant role in adipogenesis and adipokine production in human adipocytes. Int J Obes (Lond) 38: 1228-1233.
- Wen X (2013) A Phenomenon found in the Transcriptional Regulation Study on Mouse Melanocortin 3 Receptor. J Obes Weight Loss Ther 3: 202.
- Butkinaree C, Park K, Hart GW (2010) O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. BiochemBiophysActa 1800: 96-106.
- Hanover JA, Krause MW, Love DC (2012) Bittersweet memories: linking metabolism to epigenetics through O- GlcNAcylation. Nat Rev Mol Cell Biol 13: 312-321.
- Bohmann D (1990) Transcription factor phosphorylation: a link between signal transduction and the regulation of gene expression. Caner Cells 2: 337-344.
- Nunez BS, Vedeckis WV (2002) Characterization of promoter 1B in the human glucocorticoid receptor gene. Mol Cell Endocrinol 28: 191-199.
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics (2005) 21: 2933-2942.
- Hart GW, Akimoto Y (2009) The O-GlcNAc Modification. In: Essentials of Glycobiology 2nd edition. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY).
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences